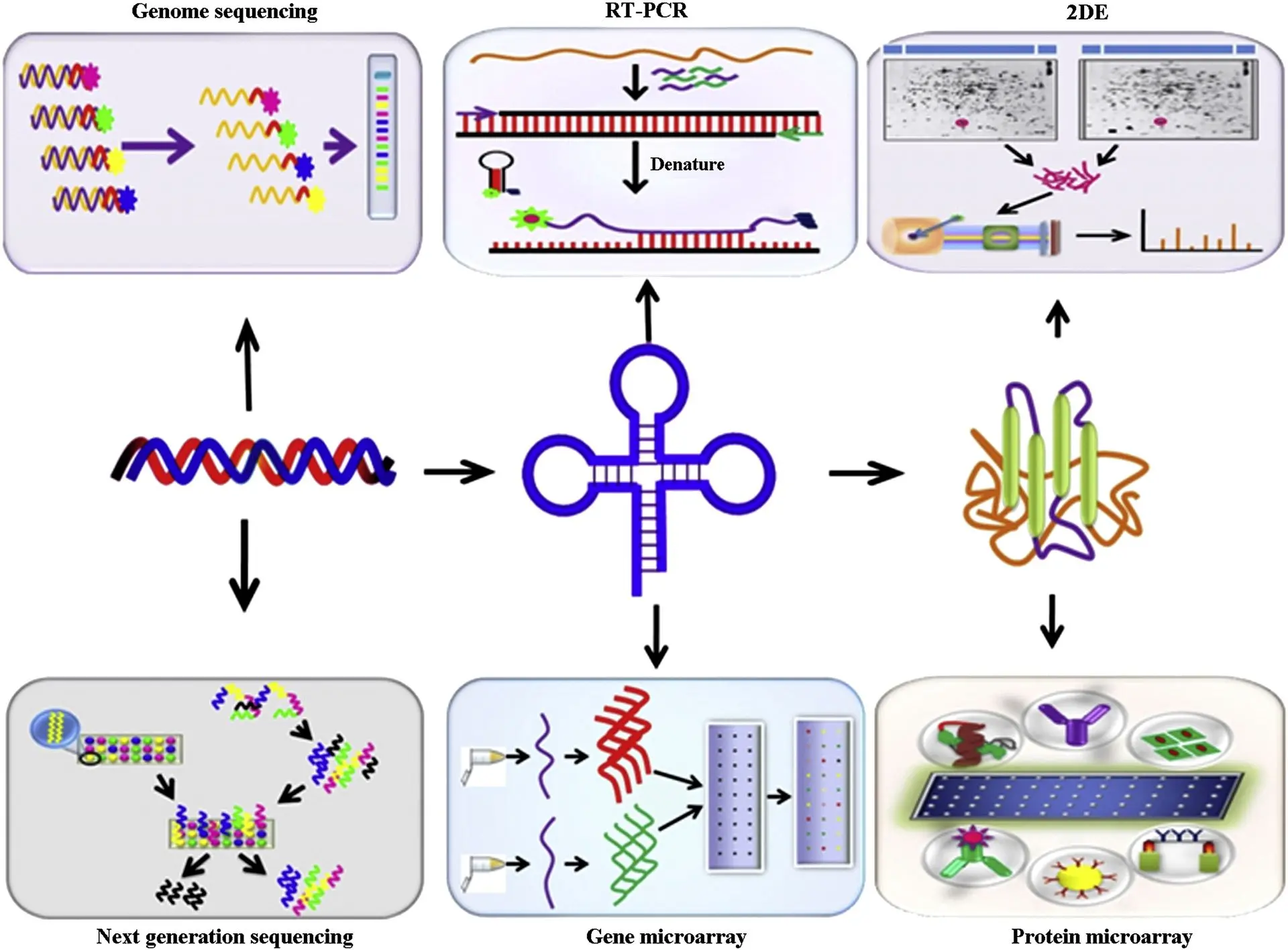
In the rapidly advancing field of proteomics, the ability to analyze protein expression, modification states, and functional dynamics across large sample cohorts has become essential to understanding complex biological systems. Whether in basic science, drug development, or clinical diagnostics, modern researchers rely on high throughput proteomic tools to dissect cellular pathways, identify disease biomarkers, and monitor therapeutic responses. At the heart of many of these technologies lies a fundamental biological tool: the antibody.
Antibodies serve as critical molecular recognition agents, enabling the specific detection of target proteins among tens of thousands of background molecules. Their ability to bind unique epitopes with high affinity and selectivity has made them indispensable in a wide range of applications including immunoblotting, immunohistochemistry, ELISA, flow cytometry, and protein microarrays. In high-throughput proteomic formats, particularly those designed to monitor protein phosphorylation or other post-translational modifications, antibodies are the key to signal fidelity and biological insight.
However, the performance of an antibody is far from guaranteed. Variables such as epitope accessibility, cross-reactivity, batch-to-batch variability, and incomplete validation can lead to significant data artifacts, misinterpretations, and reproducibility issues. These challenges are magnified in miniaturized or multiplexed assays, where antibody performance must remain consistent across hundreds or thousands of samples, often in conditions that differ from traditional immunoassays.
In response to these limitations, the antibody field is undergoing a wave of technological innovation and reinvention. From the development of recombinant monoclonal antibodies with defined genetic sequences to engineered single-domain binders like nanobodies, affimers, and aptamers, scientists now have access to a growing arsenal of next-generation affinity reagents that offer improved specificity, reproducibility, and scalability.
These advances are not only addressing long-standing challenges in antibody reliability but are also expanding the frontiers of proteomic discovery. Improved binders are enabling more accurate measurement of low-abundance proteins, finer resolution of post-translational states, and better compatibility with automated, high-throughput workflows. In clinical settings, the move toward standardized, validated, and recombinant reagents is becoming essential for regulatory compliance and assay reproducibility particularly in oncology, immunology, and neurodegenerative research.
As proteomics continues to evolve toward greater sensitivity, throughput, and clinical relevance, the quality of antibodies and their emerging alternatives will remain a defining factor in the success of these efforts. Innovations in antibody engineering, validation pipelines, and synthetic binder development are rapidly transforming how proteins are studied, compared, and understood at a systems level.
Why Antibodies Matter in Reverse Phase Protein Microarrays (RPPA)
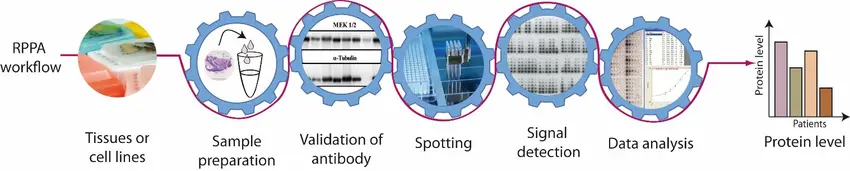
In Reverse Phase Protein Microarrays (RPPA), the role of the antibody is far more than passive detection it is the core element that defines assay performance. In this highly miniaturized and quantitative platform, protein lysates derived from cells, tissues, or clinical samples are serially diluted and meticulously arrayed onto a solid support, often nitrocellulose coated glass slides or hydrophobic polymer substrates. These microarrays can contain hundreds to thousands of samples spotted in precise configurations, enabling large scale proteomic profiling using minimal sample volumes.
Once immobilized, these arrays are probed with a single primary antibody per slide, allowing researchers to measure the expression level, activation state, or post-translational modification of a specific protein across all the samples in parallel. This one-antibody-per-slide approach provides highly quantitative, reproducible data but only if the antibody used is of exceptionally high quality.
Because RPPA is a single-binder assay (no sandwich system or signal amplification through secondary binding pairs), the burden of detection specificity and fidelity rests entirely on that primary antibody. This imposes strict requirements that go far beyond the typical validation needed for Western blot or immunohistochemistry.
1. High Specificity for Isoform and PTM Discrimination
RPPA is frequently used not only to measure total protein abundance but also to differentiate between specific isoforms and post-translationally modified states such as phosphorylated, acetylated, or cleaved variants. For example:
- In cancer research, phospho-AKT (Ser473) versus total AKT levels may signal pathway activation.
- In neurodegeneration, cleaved caspase-3 serves as a marker for apoptosis, distinct from its inactive pro-form.
Antibodies used in RPPA must recognize the correct epitope with absolute precision, avoiding cross reactivity with homologous proteins or other isoforms. This is especially critical when monitoring signaling cascades where multiple proteins share conserved motifs.
2. High Affinity for Low-Abundance Targets
Unlike highly abundant structural proteins, many biologically important proteins such as transcription factors, signaling intermediates, or phosphorylated kinases exist at low endogenous levels. Antibodies in RPPA must exhibit strong binding affinity (typically in the low nanomolar or sub-nanomolar range) to reliably detect these scarce targets.
In clinical RPPA applications, where patient derived specimens (tumor biopsies, cerebrospinal fluid, or serum) may be limited in quantity and heterogeneity, this requirement becomes even more stringent. High-affinity antibodies allow for lower background, better signal-to-noise ratios, and more accurate quantification in such complex biological matrices.
3. Batch-to-Batch Consistency and Scalability
RPPA is frequently used in longitudinal studies, clinical trials, and cohort-wide biomarker discovery projects. In these contexts, reproducibility is paramount. Any variation in antibody performance across lots or over time can lead to inconsistent results, undermining statistical power and biological interpretation.
Therefore, antibodies used in RPPA must be:
- Clonally defined (preferably recombinant),
- Available at large scale for extended use,
- Subjected to strict quality control and characterization.
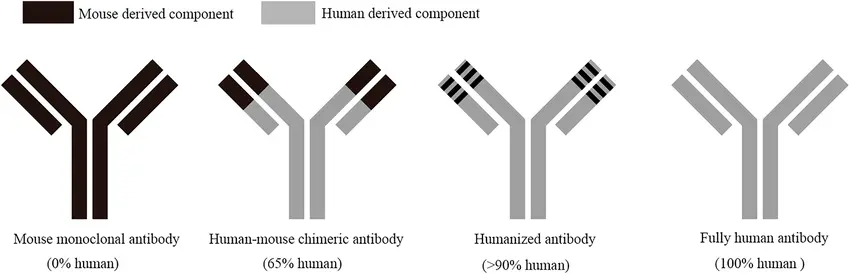
This is one of the reasons why recombinant monoclonal antibodies are increasingly favored over traditional polyclonal reagents they eliminate the variability associated with animal derived immunization and enable consistent manufacturing pipelines.
4. Minimal Cross-Reactivity and Non-Specific Binding
Given the complexity of protein lysates and the wide dynamic range of protein expression, non-specific antibody binding can lead to false positives, signal bleed, or data artifacts. RPPA requires antibodies with:
- Low background signal in denatured or diluted lysates,
- No binding to irrelevant proteins or cellular debris,
- Absence of Fc-mediated aggregation or off-target interactions.
Even weak cross-reactivity can skew quantification across hundreds of samples, especially in systems where the target protein is closely related to others (ERK1 vs. ERK2, or PD-L1 vs. PD-L2). To prevent this, cross-validation using siRNA knockdown, knockout controls, or peptide blocking is essential.
Monoclonal antibodies targeting post-translational protein modifications are essential tools for precise and quantitative proteomic analyses. Buy it from maxanim
Antibody Innovations Transforming RPPA
Recent technological advances have dramatically improved antibody performance in RPPA assays. Let’s explore the most promising trends:
1. Recombinant Monoclonal Antibodies (rAbs)
Recombinant antibodies are engineered using cloned antibody genes, ensuring unmatched:
- Batch-to-batch reproducibility,
- Scalability for clinical-grade use,
- Customizability (epitope targeting, species adaptation),
- Humanization for therapeutic translation.
2. Nanobodies and Single-Domain Antibodies
Nanobodies are small, single domain antibodies derived from camelids (alpacas). They bring several key advantages to RPPA:
- Small size (~15 kDa) allows better penetration and binding in dense microarrays,
- Exceptional stability under denaturing and high-temperature conditions,
- High specificity even for conformational epitopes.
3. Affibodies and Scaffold Proteins
Affibodies are synthetic affinity proteins based on a three-helix scaffold, engineered to bind targets with high specificity and affinity. Compared to traditional antibodies, affibodies:
- Are smaller and more stable,
- Can be easily synthesized or conjugated,
- Offer a cheaper and scalable alternative for industrial RPPA applications.
4. Aptamers: Nucleic Acid-Based Affinity Reagents
Aptamers are single-stranded DNA or RNA oligonucleotides that fold into 3D structures to bind target proteins with high affinity. In RPPA, aptamers:
- Are synthetic and reproducible,
- Can be selected against challenging or toxic proteins,
- Are easily labeled for signal detection.
5. Antibody Validation and Standardization Platforms
Reliable RPPA requires rigorous antibody validation. Today’s best practices include:
- Western blot + RPPA cross-validation
- Peptide blocking assays
- Gene knockdown/knockout controls (CRISPR-Cas9)
- Access to large antibody databases (CPTAC, Antibodypedia, CiteAb)
Conclusion: Better Antibodies = Better Proteomics
In RPPA, the antibody is not just a tool it’s the foundation. With innovations in antibody engineering, synthetic binders, and validation platforms, researchers can now conduct more reliable, reproducible, and clinically relevant proteomic studies.
The Anti AGEs Monoclonal Antibody is an essential tool for precise and quantitative analysis of post-translational modifications. Available from Maxanim.